A team of international researchers, including Dr Michel Thiebaut de Schotten our Deputy Director of Artificial Intelligence & Imaging, has succeeded in analysing brain mitochondria to create the first probabilistic map of mitochondrial biology. Read on.
Why take an interest in brain mitochondria?
The brain is the most energy-hungry organ in the body. Constituting only 2% of body weight, it eats 20-25% of the body’s energy budget. And it never stops, even when you are asleep. However, nothing is free in biology, and everything has an energy cost. That is why we breathe and eat – to give our bodies energy that fuels their functions. Likewise, thoughts, emotions, and sensations require energy transformation.
The brain’s high burn rate powers synapses and makes new proteins required for its complex functioning—from encoding our memories to sustaining the oscillations required for consciousness. To power its activities, the brain has loads of mitochondria: small bacteria-derived energy and information processors. Each brain cell contains thousands of mitochondria. They populate the cell bodies, the axons and dendrites of neurons, as well as the cytoplasm of the dozens of glial cell types that keep the brain alive.
But how many mitochondria does the brain have exactly? Are they uniformly distributed across the whole brain, or are there regions with more and others with less? Are all brain mitochondria created the same, or do they specialize in different functions depending on the brain region? For example, are mitochondria in the white matter– the myelinated highways that connect distant brain regions – different from the mitochondria in the gray matter? How about mitochondria in various cell types?
The new paper A Human Brain Map of Mitochondrial Respiratory Capacity and Diversity addressed these questions by bridging a major scale gap between the subcellular mitochondrial biology and the macro-level anatomical structures and neuroimaging models that are the focus of cognitive neuroscience.
MitoBrainMap v1.0, an unprecedented research project
The MitoBrainMap v1.0 project was directed by Dr. Martin Picard, Associate Professor and director of the Mitochondrial Psychobiology Group in the Departments of Psychiatry, Neurology, and the Butler Columbia Aging Center, who also co-directs the Columbia Science of Health program of the University of Colombia (New-York, USA).
This work is exceptional for the three following main reasons:
- The new method of brain voxelisation
- Mitochondrial profiling
- The computer model for predicting mitochondrial characteristics
The brain voxelisation method
The first significant advance is the new method invented by the authors to physically “voxelize” the frozen human brain into small cubes, or voxels 3x3x3mm in size.
Without systematically portioning massive brain slices into small pieces, it would not be possible to molecularly and enzymatically profile their mitochondria.
“Simple engineering solutions are usually the best in terms of reliability and reproducibility, and these characteristics were very important for us as we only had one shot at it”, said Dr. Eugene Mosharov, Research Scientist in the Department of Psychiatry and New York State Psychiatric Institute and first author on the paper, who designed the hardware-software solution for the problem.
His voxelization approach, which used a CNC cutter that is typically employed for wood curving and small DIY projects, allowed the team to systematically partition a frozen brain slab from a healthy 51-year-old male who died suddenly of a heart condition. Less than 8 hours later, his brain was sliced and frozen, awaiting 10 years the moment science would be ready to make good use of it. The cause of death and short postmortem delay made this brain particularly valuable to map the distribution and diversity of mitochondria in a healthy brain.
The brain was provided by the Quantitative Brain Biology (Brain QUANT) Institute in the Department of Psychiatry, directed by Maura Boldrini. For over 20 years now, Gorazd Rosoklija and Andrew Dwork have curated hundreds of precious human brains. With their collaborator John Mann, they perform “psychological autopsies” on each donor, ascertaining their mental health status. An exceptional resource for neuroscience research.
Mitochondrial profiling
The second advance is to molecularly define the bioenergetic landscape of mitochondrial phenotypes and diversity across brain regions.
This includes white matter and different portions of the cortical and sub-cortical gray matter regions, which the team showed have qualitatively distinct mitochondria density and oxidative phosphorylation (OxPhos) capacity. Quite surprisingly, the authors also found a correlation between the mitochondrial capacity to transform energy and the estimated phylogenic age of brain regions, with higher OxPhos capacity in brain areas that appeared later in evolution.
The mitochondrial profiling was performed in collaboration with two other teams. Phil De Jager and Ya Zhang at the Center for Translational and Computational Neuroimmunology of the University of Colombia, who performed single-nucleus RNA sequencing with Anna Monzel in Dr. Picard’s laboratory. And Orian Shirihai’s team including Linsey Stiles and Corey Osto at the UCLA Metabolism Theme, who deployed a frozen tissue respirometry technique they pioneered a few years back. It was the first time they performed their assays on hundreds of samples from a human brain.
“Across four voxels from different brain regions, the single-nucleus gene expression data showed us the expected proportions of cell types, and even that mitochondria were qualitatively different”, remarked Monzel, a senior computational scientist in Dr. Picard’s group. “Different cell types and brain regions have different types of mitochondria, or mitotypes.”
The computer model for predicting mitochondrial characteristics
The third advance came from a computational model extending the data from a single brain slice to the whole brain.
Dr. Michel Thiebaut de Shotten, VBHI deputy director for AI and Imaging and Research Director at CNRS, transposed the mitochondrial data from this brain section to the “standard” brain in neuroimaging research.
This then allowed the team to achieve several advances:
- The first was to visualize the biochemical data in the same reference space as every human neuroimaging research is typically done.
“This makes the mitochondria relatable to other modalities” explains Picard. In this way, the novel mitochondrial results can be visualized and interpreted in relation to decades of past research.
- The second was to build a fairly simple computational model to predict molecular markers of mitochondrial density and energy transformation capacity from neuroimaging modalities.
“Based on a few simple MRI metrics that most laboratories can acquire, and that many datasets already contain, in and out of sample test we were able to predict mitochondrial features with a decent 35-40% accuracy”, said Dr. Thiebaut de Schotten.
The team found that standard structural and functional neuroimaging parameters contained sufficient information to predict some aspects of mitochondrial biology.
“If true, this could mean that something in the biophysical properties of mitochondria is embedded in standard magnetic resonance-based signals”, remarked Picard.
- The final step consisted in expanding the prediction model to the scale of the whole brain.
This led to the first probabilistic map of mitochondrial biology, meaning that even though we never measured mitochondrial features in some deep part of the temporal lobe, our model can predict the amount and types of mitochondria found there based on MRI data.
Further research is now needed to test, validate and apply this neuroimaging-based mitochondrial profiling approach in various biological and clinical contexts.
“If this works, it would be the first non-invasive way of peering into the biology of mitochondrial bioenergetics in the living human brain” said Dr. Thiebaut de Schotten.
“This is the kind of project that is nearly impossible to do” said Picard, “we had to find an exceptionally high-quality brain, align engineering, computational, molecular biology, and neuroscience expertise, and find the right students courageous enough to undertake this massive effort!”
The courageous students were a pair of recent graduates, Ayelet Rosenberg and Snehal Bindra, who performed what is likely the largest mitochondrial biochemistry project to date.
“I am so grateful for this amazing team that came together to create MitoBrainMap v1.0”, said Snehal Bindra.
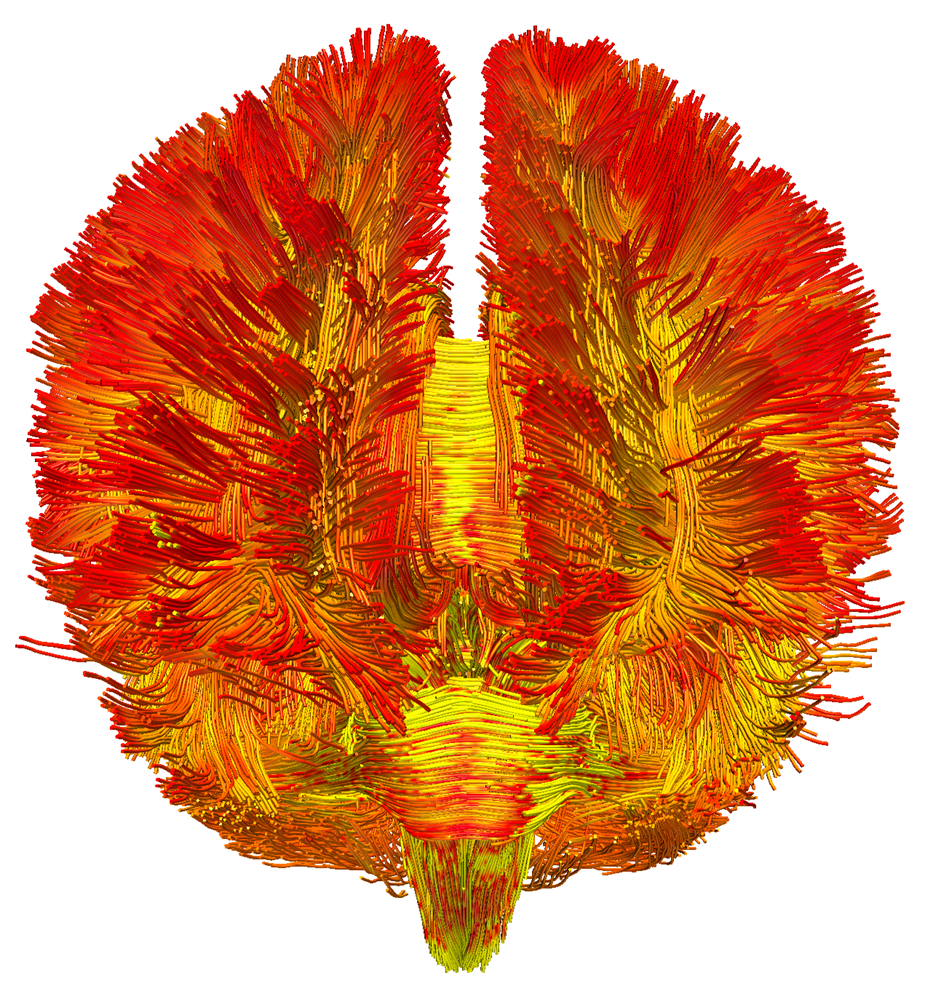
Picture caption: The first 3D map of mitochondrial density in the living human brain, visualised on the structural connectome. This innovative illustration integrates mitochondrial density (the redder the colours, the higher the density) with the organisation of neuronal fibres, providing a complete view of grey/white matter differentiation and mapping on a brain scale. ©Michel Thiebaut de Schotten
The brain voxelization method and the brain mitochondrial findings in this landmark paper reveal the landscape of human brain mitochondrial energetics.
It provides a new tool to understand how brain energy may relate to anatomy, development, behavior, neurodegeneration, well-being, and perhaps even biggest questions related to consciousness.
Scientific publication
Mosharov, E.V., Rosenberg, A.M., Monzel, A.S. et al. A human brain map of mitochondrial respiratory capacity and diversity. Nature (2025). https://doi.org/10.1038/s41586-025-08740-6
Find out more information and behind the scenes pictures on http://humanmitobrainmap.bcblab.com